chi-squared and linkage
Chi-square
Here is a short ch-square problem
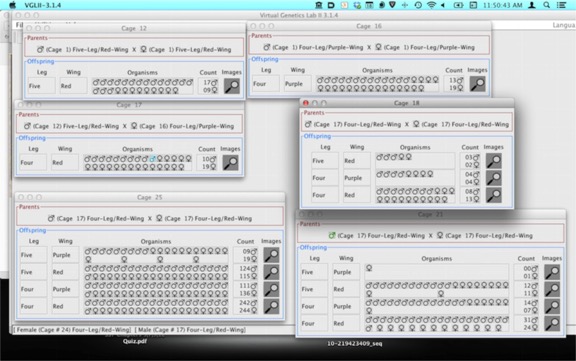
This is a Dihybrid cross, meaning AaBb x AaBa. That means the expected results are 9:3:3:1. That won’t be what you get.
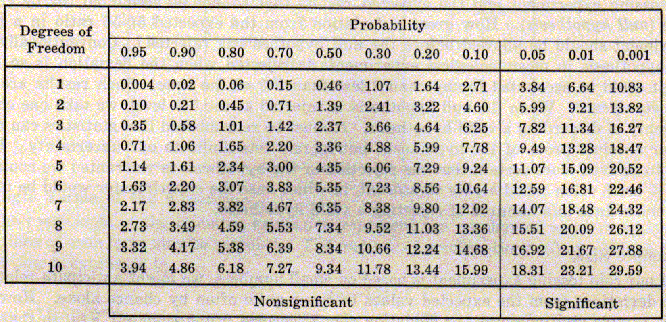
Cage 17 is the F1 dihybrid. Cages 12 and 16 are there because the were the source of the P1 generation.The remaining three cages, 18, 21 and 25 are just increasing numbers of offspring. Try performing a chi-square for 18, 21 and 25. It’s pretty striking how the size of the data set makes such a difference. There is a chi-square table below to assess the probabilities.
A few questions you should be able to answer: What are the “Parental” types you expect to see more frequently if the genes are linked. Do you think the genes are linked? Explain your reasoning.
Pedigree
Here is a link to a site at Stanford that helps you understand pedigree. If the link downloads a file instead of opening a page, you have to do the following: right-click or control-click the link and select "copy link." Then paste it into an address line and change the ending from "swf" to "html." Then it should work.
There is a short quiz you can take. I won’t see the results.
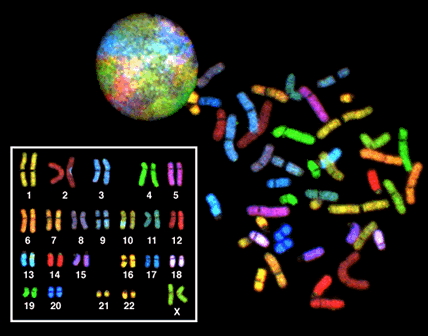
and here is the Karyotype activity
Below is a blog I wrote on the topics. You can use it as a reference if it helps.
Karyotyping
:(as usual, images from wikipedia).
Karyotyping is a technique for looking at all the chromosomes in a cell. Briefly, a pro-metaphase cell is is stained with a dye that binds chromosomes. There the traditional dye, giemsa, stains the chromosomes in a banding pattern that is different for each one. So, the banding pattern of chromosome 1 is different from that of chromosome 2, etc. These days, there are specific color-coded probes that make it easier. The cell is then photographed and the picture is enlarged. Then, traditionally by literally cutting up the picture, but now digitally, the chromosomes are cut out and aligned so that we can see that the individual has 2 copies (one from mom and one from dad) of each of the 22 autosomes. The largest is chromosome 1 and the smallest is chromosome 22.
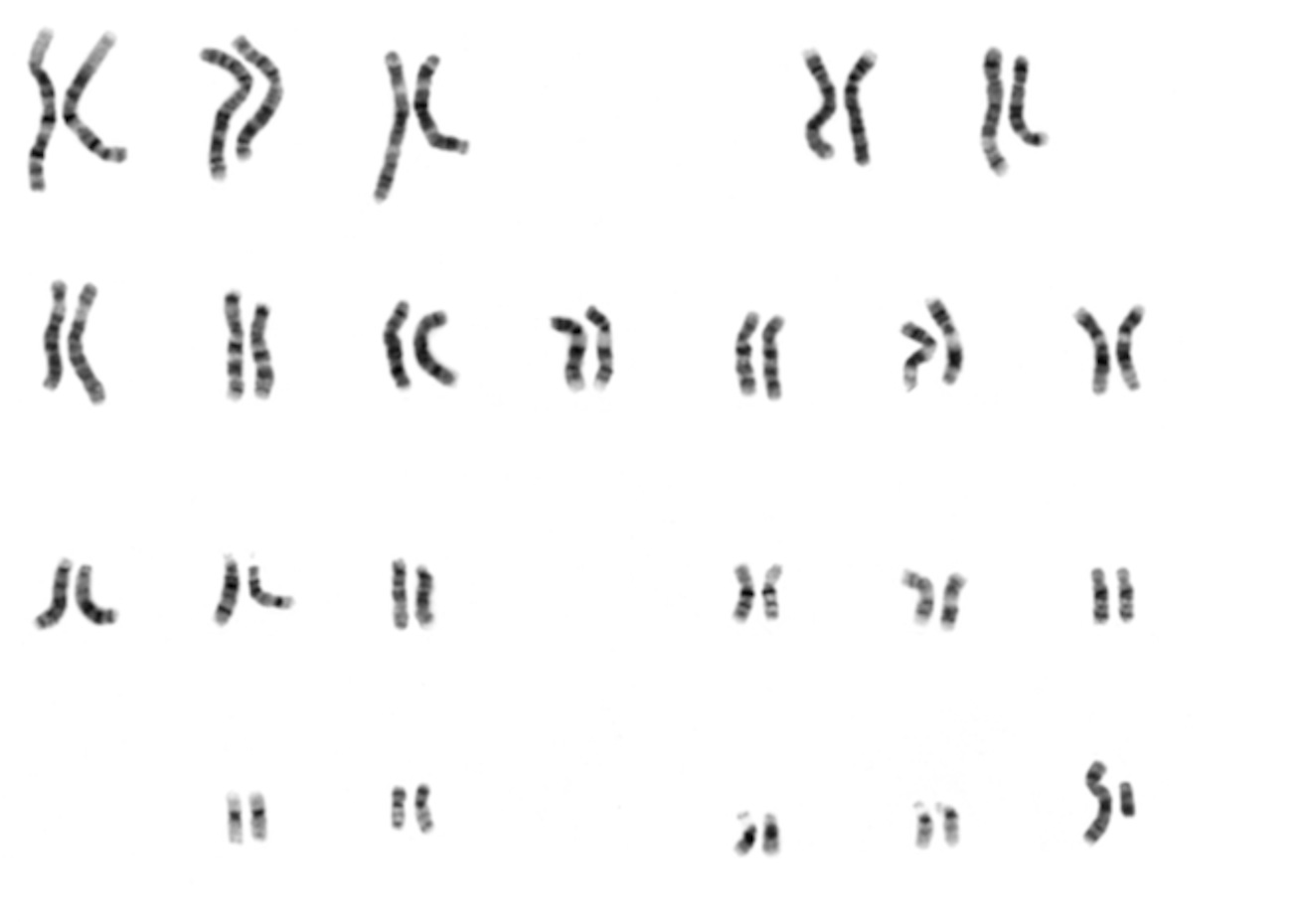
Then look at the last pair: The really tiny one, smaller even than chromosome 22, is the “Y” chromosome (so this is a male). The larger one next to it is the X.
Look how much cooler it is with the modern technique:
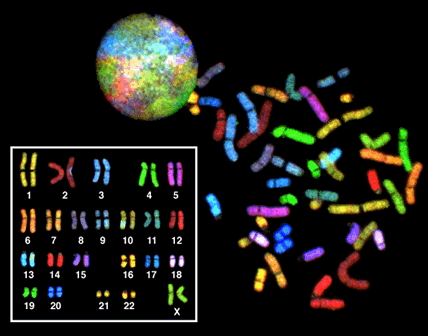
As you can see, this individual is a female.
Using techniques such as this, we can look for large changes to the DNA, rearrangements of the chromosomes we call “inversions,” “deletions” (both self explanatory), translocations (where a piece of one chromosome is spliced to another), extra or missing copies of one chromosome (“trisomy” or “monosomy”) or, even whole extra sets of chromosomes.
Some common disease states result from this. Trisomy 21 causes Down syndrome.
This can occur when the sister chromosomes of 21 do not separate at meiosis I (this is called “nondisjunction”). That results in a 2 gametes getting no chromosome 21 and 2 others getting 2 copies of it. If one of that latter group fuses with a normal gamete, the resulting zygote has 3 copies.
Check out this karyotype:
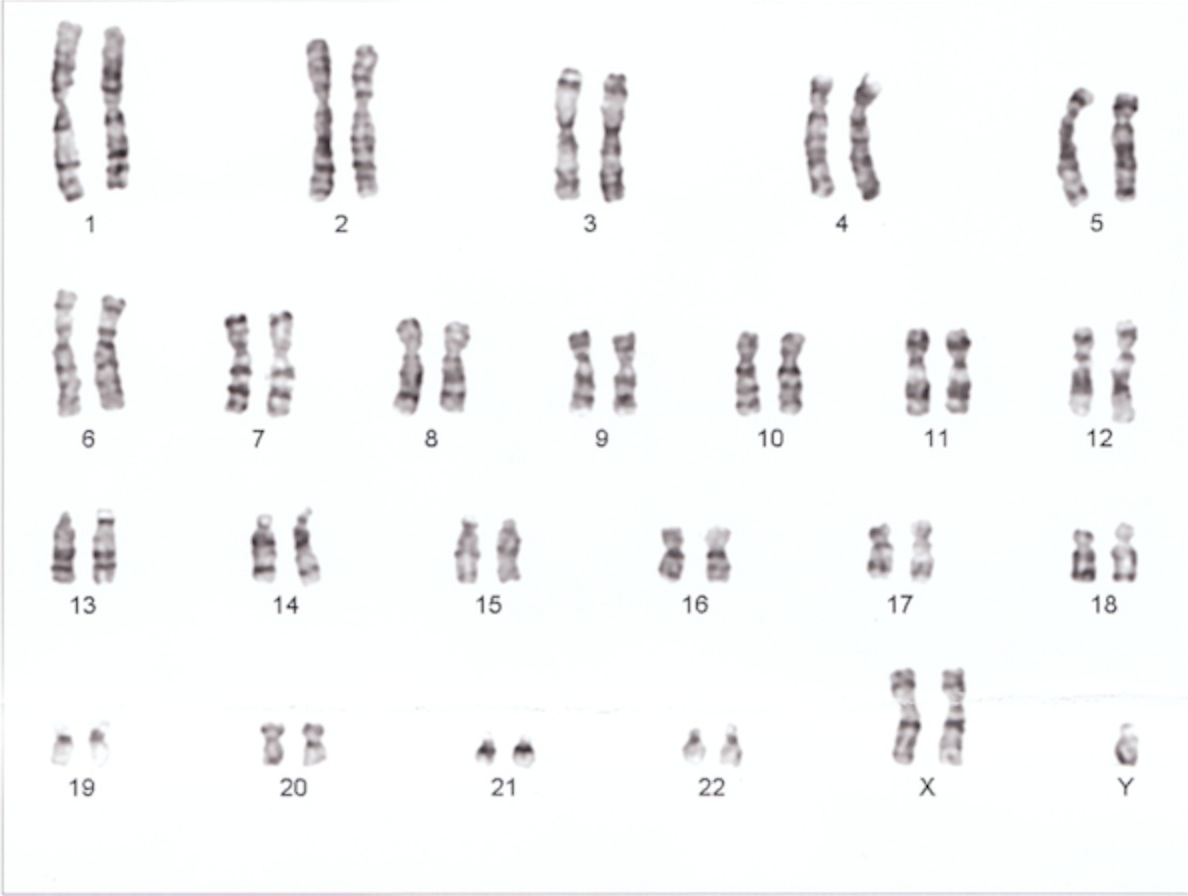
Do you see what’s wrong? This person has Kleinfelter’s syndrome, where he has 2 X chromosome and a Y.
Pedigree Analysis:
Here is the pedigree example given in the Campbell Book.
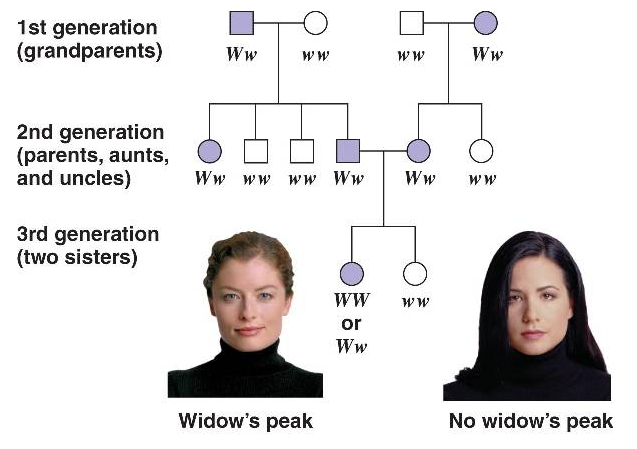
Females are circles, males are squares. People with the trait being followed usually are filled in with a color, people without the trait are not. This trait, the appearance of the “widow’s peak,” is a dominant allele on one of the Autosomes (not sex linked). How can I tell? Remember that the recessive allele can be covered up by a dominant allele. So, two people with the trait, if it is dominant, can produce a child without it. Notice that this happens in the mating in the second generation. Two “widow’s peak” people had a non widow’s peak child. You never see enough children to invoke statistical predictions in a generation. You should see only ¼ homozygous recessive in the 3rd generation…but there are only two kids. Even if there were 4, the numbers are not good enough to make predictions.
I know it is not on the X because an affected mother in generation 1 married an unaffected man and had an affected daughter. That would be impossible for a recessive allele on the X (the daughter would have to have 2 copies, one from each parent. But Dad cannot have it since he does not show the trait). I know it is not on the Y because it appears in women.
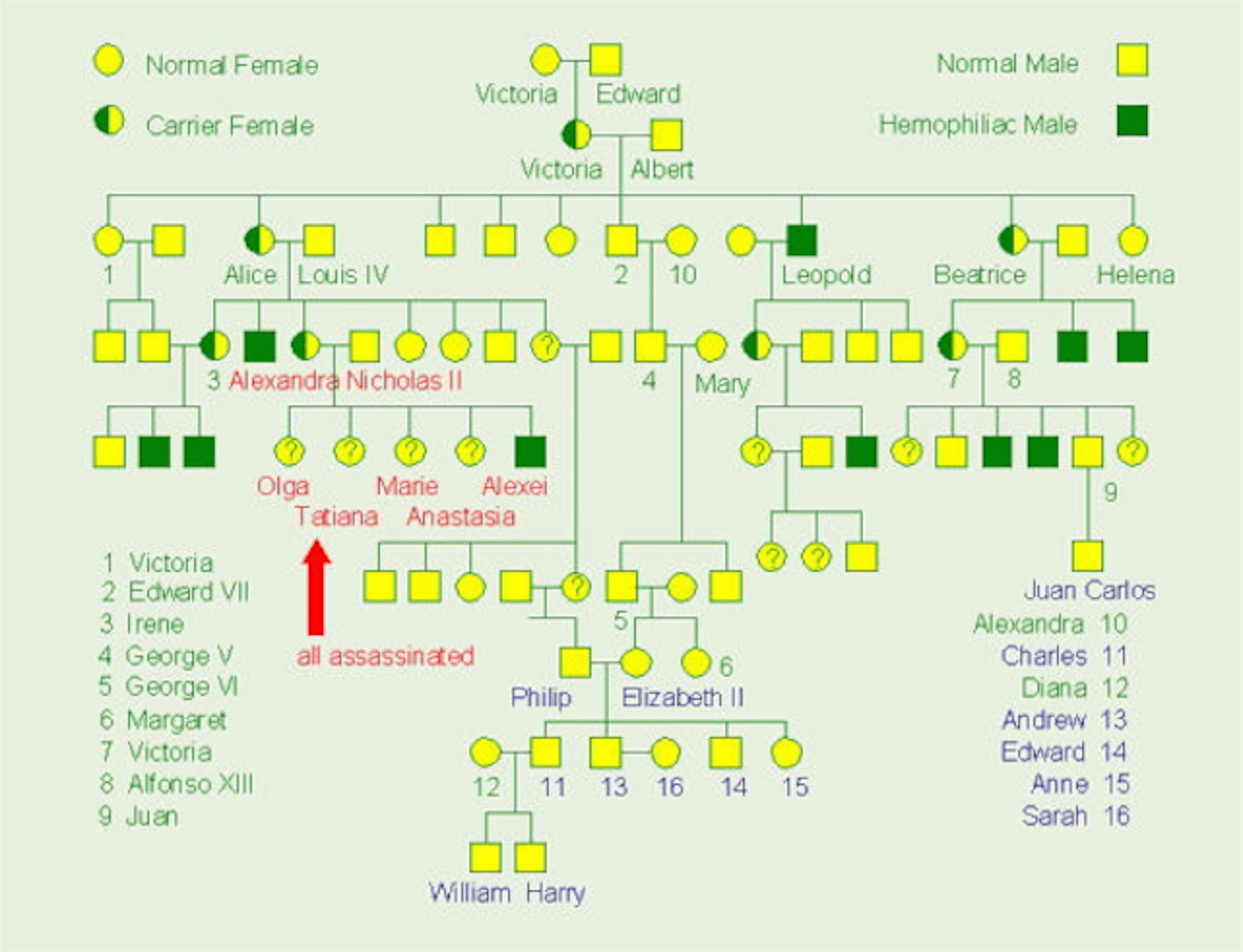
Here is the pedigree for hemophilia (a severe bleeding disease) in the royal families of Europe. The mutation seems to have occurred in Queen Victoria.
- Is this trait sex-linked?
- How do you know?
- How do we know that Irene (number “3”) is a carrier?
- Is it possible that Queen Elizabeth is a carrier?
- Could the disease show up in any of the children Prince William and Kate Middleton might have?