Dihybrid crosses
Diploid Crosses
Single Locus
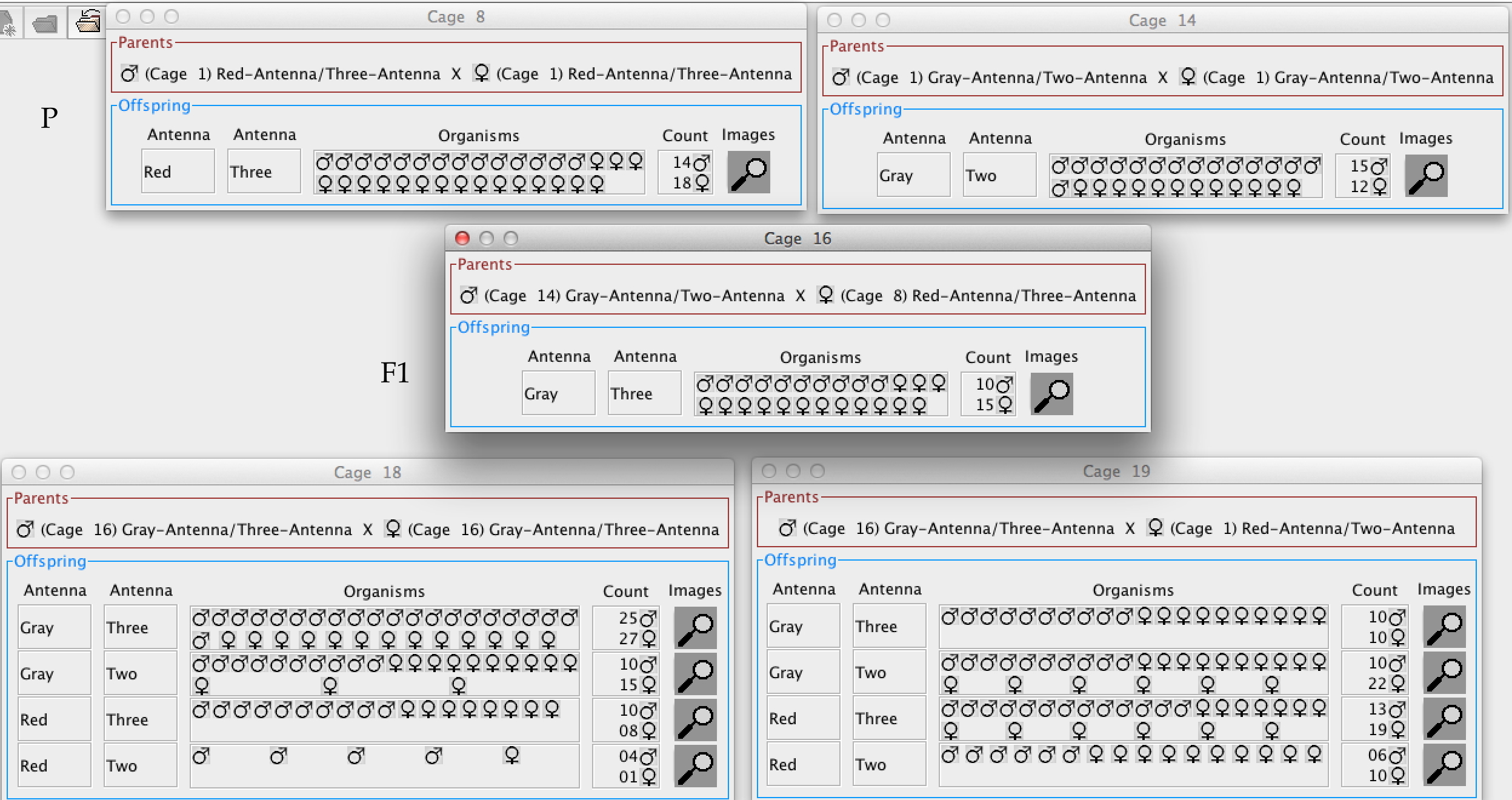
As we've learned, if there are two alleles at one locus not on sex chromosome, the hybrid cross (Aa x Aa) must produce the 1:2:1 assortment. However, the 3:1 dominance is only predicted if there is a simple dominance relationship. aa x aa is boring. The other three types of crosses are shown below, AA x aa; Aa x aa and Aa x Aa

Sex linked
Here's how that maps out for a simple dominance on the X chromosome:
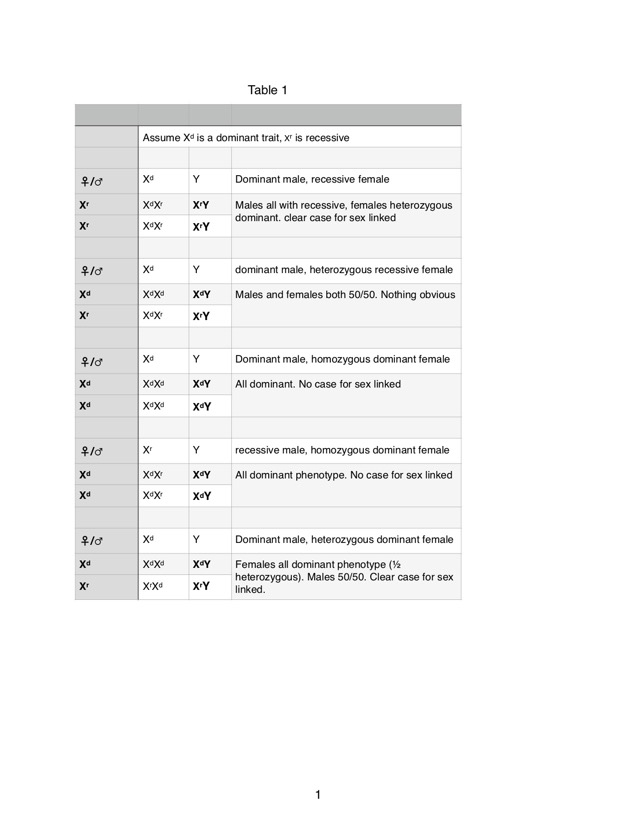
For incomplete dominance, the case is interesting because the heterozygous phenotype ONLY shows up in females.
Multiple Loci
The first think is how do we deal with multiple loci? There is a way that is done purely with math…that's what we do when there are more than two loci. However, I'm going to show you how to do it with a punnet square.
You can download a plot of the possible gametes and that 4x4 matrix here. It is also shown below.
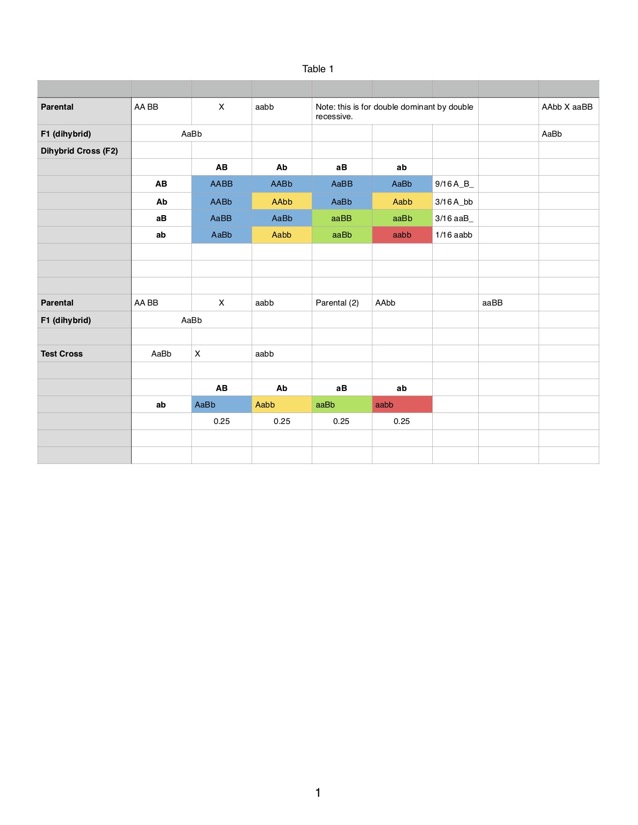
The expected 9:3:3:1 ratio assumes independent assortment (not on the same chromosome). We can use that prediction to ask whether there is linkage (are they really on the same chromosome) and test the model out using a statistical test we will cover next.
Some variants in inheritance.
Multi-gene trait. There's really not much to this. Some traits…really most interesting traits…may be inherited, but be based on more than one gene. So, there is no single gene for being tall. There is no allele for being 6' tall. But, that doesn't mean that height is not inherited. Many different loci may contribute to it. Here is a hypothetical plot of what you might expect if there are three loci that contribute to height, each with two alleles where the dominant of each allele contributes positively to height (note, there is no reason any of those assumptions should be true). You might see a distribution that looks something like this:
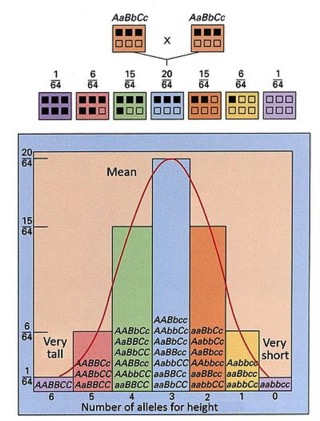
So, you can get a distribution of heights based on some combination of alleles at different loci. In this case, we are assuming simple additive interactions among the genes. It an be more complex (See below(.
Pleiotropy
This is sort of the opposite of multi-gene trait. Here, one gene can affect many traits. We've discussed this in the context of cytoskeletal proteins before. For example, mutations to a microtubule-associated motor protein could affect male fertility (sperm flagellum), airway function (cilia on airway epithelium) and vesicle transport and secretion of proteins. Everywhere that protein is needed, you would see some effect.
Epistasis
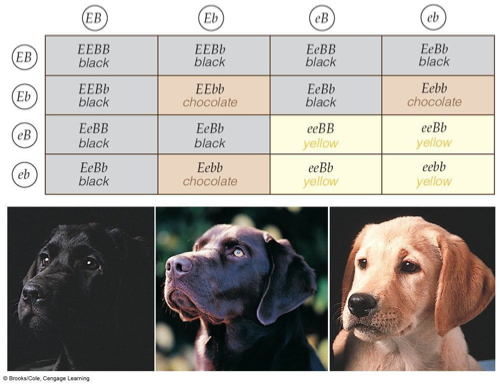
Finally, there is epistasis. Proteins interact with other proteins, so variations in one gene can affect how you see the phenotype caused by another. Here is a classic example I stole from another website at the university of Georgia. It covers coat color in.
Labrador retrievers. One locus, the B locus, controls the color of the pigment eumelanin. Eumelanin can be either brown in color (bb) or black BB or Bb.
Another gene, known as the "E" locus (for extensor…never mind) is needed to deposit eumelanin in the fur of the dog. It encodes a protein called MC1R and where it is expressed determines whether the eumelanin gets into the fur. The ee homozygote does not deposit eumelanin at all in the fur while Ee or EE do.
Thus, if the dog is ee, it will be yellow no matter whether it makes black or brown melanin. All the possibilities on the 4x4 matrix are shown below.