Telomeres and DNA Replication
DNA Replication
Let me lay out the basics:
Since DNA is two complimentary strands, each strand contains the information to specify the other. Thus, it seemed logical from the first time the structure was determined that the two strands would separate and new subunits (deoxyribonucleotides) would be added to make each new strand, using the other old strand as a template. Each new double helix would therefore really be one old strand and one new one. Here is the basic idea in video. Like most of the videos I will link, these come from the Howard Hughes Medical Institutes (HHMI). Note that this video shows the new strands being made the same for both templates. As we know (and the video alludes), this cannot happen.
The first problem:
So, one strand is the template for the other. New bases are added one at a time via a simple chemical reaction we have talked about, mediated by a complicated enzyme machine (comprising many different proteins).
The problem is that the chemistry requires that a new subunit can only be added to the 3’ end. So, if you are moving along a replication fork, one strand cannot be replicated easily…the fork is moving the wrong way and it has to be replicated “backward.”
Here’s a more basic video that shows you how an Origin of replication might work and some detail, but in a much simpler form.
Here are two other videos here and here that have merit, though all of them, including the cool one below, have errors in them.
Notice that there is a second problem. As the video says, you need a short RNA primer to begin each section when synthesizing the lagging strand. This is put down by an enzyme called “primase.” The leading strand needed an RNA primer to get started too. But, since it is replicated continuously, it only needs one primer, way back at the start of replication.
Here is a link to the really cool video. I think you should look at it again, now that you have seen the simple one. We have to name all the enzymes and talk more about details tomorrow.
Here are the details of the problem
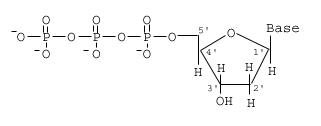
The unit of DNA polymerization (Synthesis) is a deoxyribonucleoside triphosphate. In the image, the “Base” would be either A, T, C or G, depending on what was on the template strand. Just like ATP, these molecules have high-energy (unstable, that is) bonds joining the phosphates. This can therefore be used in transfer reactions, just like enzymes transfer phosphates from ATP in reactions we have studied. There is a seemingly subtle change: instead of the third (𝛄 or “gamma”) phosphate on the end being attacked by the OH on the 3' carbon, the first one is (called “α”). This change has a big impact, though. It links the 3' carbon of the existing DNA to the 5' carbon of the incoming base via a phosphate.
This is called a “phosphodiester.”
Here is a specific example, deoxyATP
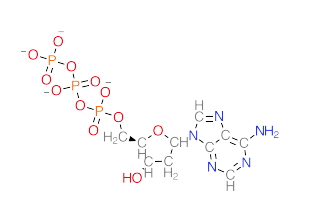
The next base that comes in will use the high-energy triphosphate it carries to attack the 3' OH.
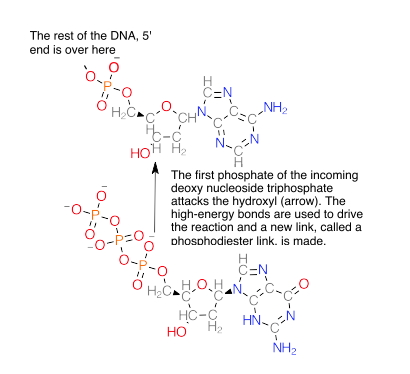
And forms this:
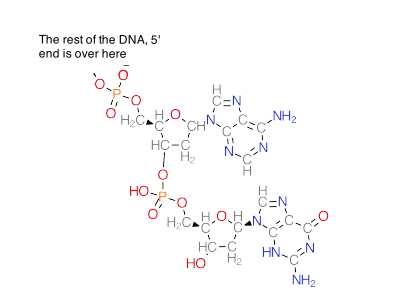
Thus, as we said, we must add DNA to the growing 3' end. From a chemical standpoint, there isn’t any reason why you cannot add an incoming base to the 5' end, provided that end has a 5' triphosphate. That triphosphate would be unstable, however. Should it hydrolyze, the new DNA wouldn’t be made until another enzyme came in and “recharged it” with new phosphates.
Telomeres and Telomerase:
After reading this, please read this short news piece from the journal "Nature."
http://www.nature.com/news/2010/101128/full/news.2010.635.html
Two problems:
- As we have discussed, the chemistry of replication leaves the lagging strand not fully replicated. There is a 3' extension of what had been the template. This would lead to shortening of the chromosome with each round of replication.
- We have not previously discussed this, but the ends of broken chromosomes, or just free ends of linear DNA, lead to recombination or “splicing” together of these fragments. Also, free ends of DNA tend to be degraded by enzymes quickly. So, what is special about the ends of our chromosomes that keeps this from happening?
Cancer and the Hayflick Limit
Our cells seem to go through a fixed number of divisions before they no longer can keep going. This is called the “Hayflick Limit” after the person who noticed it.
However, cancer cells are immortal…they overcome this limit. Also, there must be a way for cells to fix the problem so that we can “reset” the Hayflick limit when a new embryo is formed. One more thing: one-celled organisms don’t have a Hayflick limit. They can grow indefinitely. All of this pointed to some enzyme activity needed to maintain the ends.
The Sequence:
Well, since we are talking about DNA, it seems likely that there is some specific sequence that corresponds to the Telomere. There is. In humans and other vertebrates, the sequence is the short repeat 5'(TTAGGG)n, where “n” is an integer between 300 and 8000. So, that can be 50,000 bases.
- How does it get there?
- How does it achieve the goals above?
Telomerase:
The enzyme Telomerase is a little unusual. It has a DNA polymerase activity and needs a 3' end to which it adds. But, it carries a short RNA that acts as its template. So, it finds the free 3' end, which is already longer than the newly replicated strand (due to the lagging-strand problem), uses the internal template to extend the repeated sequence over and over again. Because it uses RNA as a template, but polymerizes DNA, it is known as an RNA-dependent DNA polymerase, also known as “reverse transcriptase,” since it is the opposite of transcription. The image is stolen from Wikipedia and shows the repeating sequence
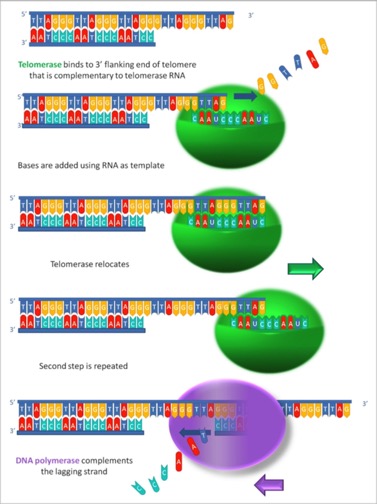
That’s how it gets there.
It fixes the shortening problem by just adding the sequence. This can then serve as a template for more lagging-strand synthesis. That will still leave a short 3' extension, but will re-lengthen the telomere.
It’s an interesting enzyme.I’ve found many other images with great detail on it, but decided to steal this one:
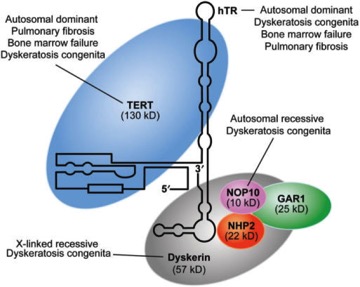
It shows that there is a long complex RNA that interacts with several proteins (Dyskerin, TERT and some smaller ones). The RNA is much more than the template. It folds into a complex structure that fits into the proteins. The reason I included this one because it makes reference to a couple of forms of a genetic disease known as “Dyskeratosis.” This is a version of premature aging where the effects are seen as premature organ failure, rather than an old-looking outward appearance. Does it make sense that mutations to genes in the telomerase complex would lead to that?
As for how it solves problem number two, the extended 3’ end will fold back around and, along with some accessory proteins, will form a complex, stable structure shown below in cartoon form:
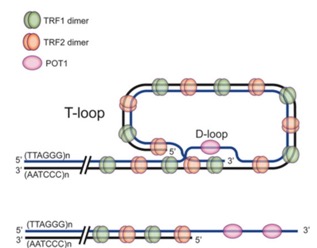
This cartoon is from from:
http://www.bioscience.org/2008/v13/af/2825/fulltext.asp?bframe=figures.htm&doi=yes):
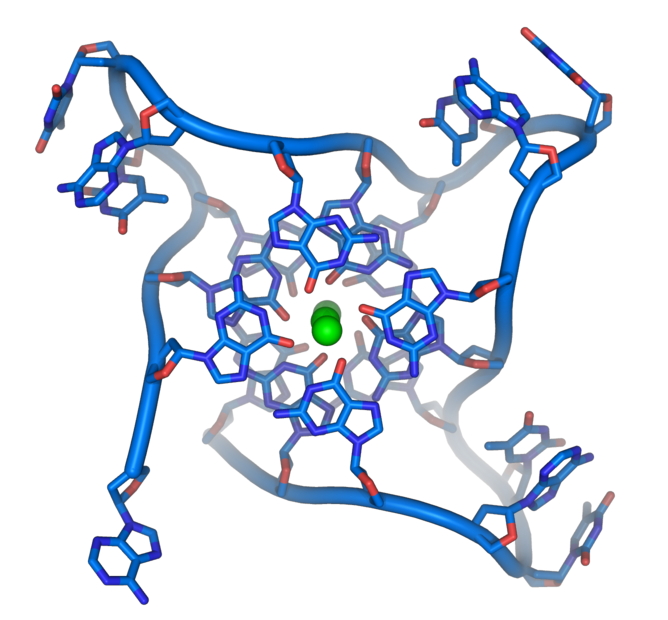
A more realistic view of the DNA in it’s four-strand form is below: (Image from Wikipedia):
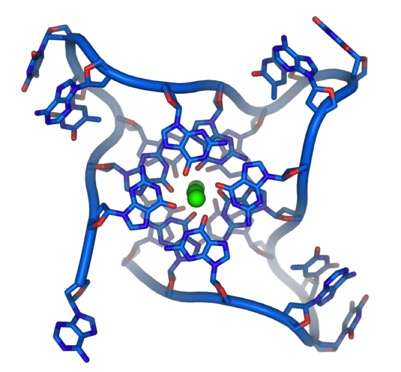
The green dot is a monvalent cation.
This depicts how the structure forms. Note that the strands are Parallel, not antiparallel.
(image from this website: http://proj1.sinica.edu.tw/~tigpcbmb/course%20material/cb9903/cb9903.htm)