Transport Proteins
Gradients
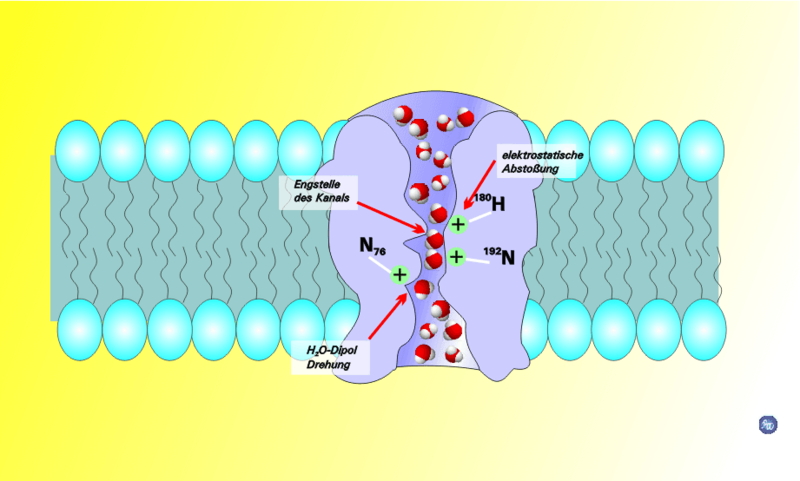
As we said, the membrane is impermeable to most dissolved solutes. Gases can pass freely through the membrane, so oxygen and CO2 can be traded as necessary. But things dissolved in the water that surrounds the cell and is inside the cell generally are hydrophilic. As such, they cannot pass through the membrane directly.
For this reason, there are transport proteins that are responsible for moving all of the small molecules, ions and even fairly large molecules across the membrane. We will talk a little bit later about the protein that allows water to move across the membrane. For right now we will talk about transport in terms of several overlapping concepts shown in the diagram.
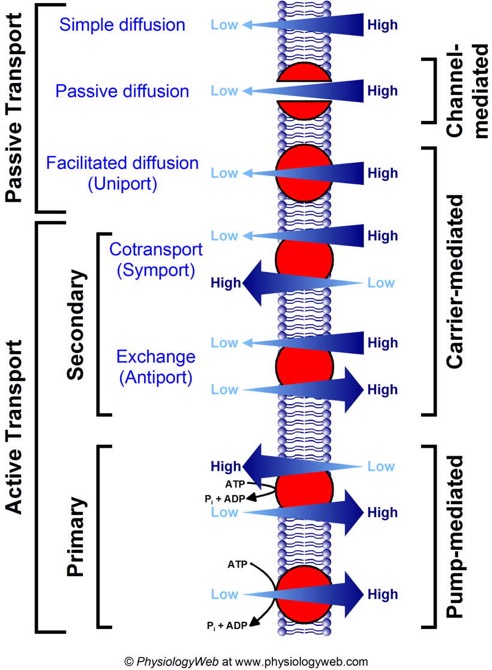
Image from www.physiologyweb.com
all forms of passive diffusion are "down gradient." That is to say, one side of the membrane has more of the solute while the other side has less. Given any pathway through which the solute can move it will tend to distribute more evenly. I can divide this further into simple diffusion for molecules that can diffuse again across the membrane directly, passive diffusion for molecules that pass through a channel protein (which is just what it sounds like, a protein that forms a channel through the membrane), and facilitated diffusion, where the solute is carried by a protein from one side of the membrane to the other, sort of like a shuttle. The important thing to remember is that all three of these are diffusion down gradient and require no input energy. No work needs to be done for molecules to diffuse. In fact as we will see later, a gradient can be used to do work.
All forms of active transport require the expenditure of energy at some point in the overall pathway and result in even greater disparities in concentration between the two sides of the membrane. That is, something moves from an area where there is a low concentration to an area where there is already a high concentration of that solute.
Active transport can be separated further into "pumps," which directly expend energy in a cycle to move solutes from one side of the membrane to the other, and "co-transport." Co-transport uses a gradient that was established by a pump or exists for some other reason to drive transport of some other molecule up its own gradient. This is one of those things that is hard to explain in writing. However the diagram below does a pretty good job of showing it.
Symport is when the molecule that is moving down gradient is moving in the same direction as the molecule you want to move up gradient.
This image from your textbook, Campbell Biology ninth edition, and shoes how a proton pump can be used to establish a gradient, then the proton gradient can be used to drive transport of sucrose "up-gradient." This is a symport protien.
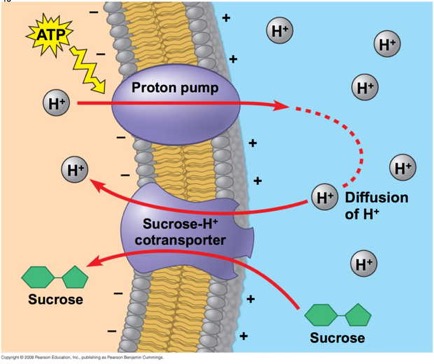
In the diagram, we see that ATP is expended to pump protons to the external side of the cell membrane.Those protons, given a pathway, would diffuse down their gradient back into the cell. The trick here is to use a pathway (that is to say, a protein in the membrane) that will only allow protons to diffuse down gradient if a sucrose molecule accompanies it.
You can think of it sort of as a revolving door that will allow protons to enter but will only revolve if sucrose is there also.
The other related concept is the "antiporter." you can think of this as a revolving door where the two players have to enter opposite sides in order for the door to turn. In either case one molecule or ion diffuses down its gradient (passive diffusion) while the other molecule moves up its gradient (active transport).
In order to carry out co-transport, one needs to have a gradient established of one or more ions. This is generally achieved by proteins that we call "pumps." While there are many that are crucial to cell function, one has an outsized role in establishing what we call the "electrochemical gradient."That protein is the sodium-potassium pump. Sometimes you will hear it called the sodium-potassium antiporter ( which makes sense because sodium and potassium ions are moving in opposite directions – but is misleading since both will move up gradient against the direction favored by diffusion.) Another name you may hear is the sodium/potassium ATPase.
Here is a moderately amusing GIF based on Drake's music video for "Hotline Bling" from a couple years ago.
Here is the gist of it: the pump can bind two potassium ions when it is open to the outside, but cannot bind sodium ions at all. When open to the inside it will no longer bind potassium ions, but can bind three sodium ions. ATP is hydrolyzed to force the switch between the two states (open to the inside versus open to the outside). The net result of this is that as it goes through a cycle two potassium ions are pumped into the cell and three sodium ions are pumped out. This results in a gradient of potassium (high on the inside, low on the outside) and sodium ions (high on the outside, low on the inside). This is the "chemical" part of the gradient. Because two positive ions are pumped in for every three that are pumped out, there is also an electrical component to the gradient. The inside of the cell ends up being negatively charged overall to a level of about -70 mV.
I would like you to read David G’s really nice blog on the pump. I think you will find some of the numbers, frankly, shocking. Also, understanding the way this works will help you with other proteins we consider.
For example, some may be interested in proton pumps in the cells of the stomach that make the stomach acidic...it works like this protein, as David explains.
Proceeding from specific to general:
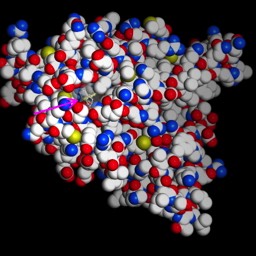
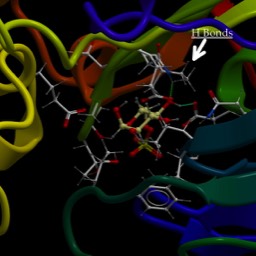
This protein has binding sites for ions. Below is an image of two ions bound. In this case, the protein crystallographers have used a trick and bound Rubidium+ to the sites. But, it works similarly. What would bind a positive ion? Well, you should hold it between negative ions, like deprotonated acid side chains. What are the acid side chains in amino acids?: Aspartic Acid (D) and Glutamic Acid (E). Look at the image below:
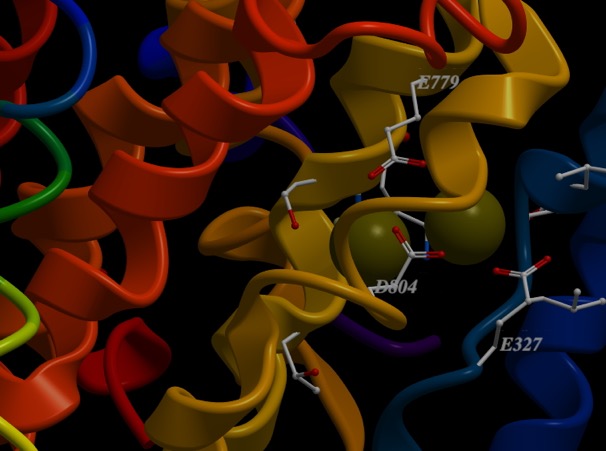
D804, E327 and E779. The numbers correspond to the residue number in the amino acid sequence, or primary structure. So, D804 is an aspartic acid that is the 804th amino acid residue in the chain and is part of one of the helices that span the membrane. E327 is a glutamic acid at position 327, and is in a different helix spanning the membrane, but resting close to D804 in the structure. All of come together to make a really nice pocket that binds the ions (greenish). That makes sense. There are others I didn’t label. All three residues that bind the cation are on different helices. If you slide those helices relative to each other, the components of the binding site move closer or farther away from each other. Now, check out the ionic radii of Na+ and K+ at the wikipedia page. K+ is 30% bigger than Na+ (Rb+ is only slightly larger than K+). So, by sliding the helices, I can change the sized of the pocket and whether K+ or Na+ is favored.
As Goodsell points out, this same mechanism can be used to pump protons. You just have to change the size of the binding pocket. That probably just involves some shifting of the positions of the acidic residues.
Gradients are useful
Once you establish a gradient, you can use it to do stuff. The pump establishes an electrochemical gradient. Every cell at “resting potential” has more K+ on the inside and more Na+ on the outside. The inside is also more negative. Maybe I could use that gradient to do something.
For example:
Phosphorylation and interactions with other molecules change the shape of proteins. Changes in the shape of proteins changes their function.
Any protein could have a set of possible shapes, each favored under different conditions, and each favored interaction in turn altering the behavior of the protein.
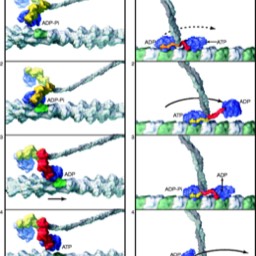
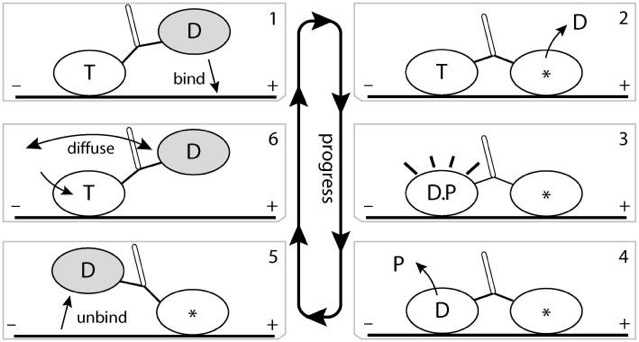
Imagine a protein that binds to microtubules, for example. Maybe you could come up with a series of shapes that change depending on whether ATP is bound, or ADP is bound, or the protein is bound to tubulin or not. Suppose you had large protein with two heads, each of which can go through a series of steps. Could you imagine a series of steps like: ATP bound to training head, ADP bound to leading head, which binds to tubular, which triggers release of ADP to the solution; which triggers the trailing head to hydrolyze ATP and release phosphate, which triggers the release of the trailing head from tubular, which triggers new ATP to bind to trailing head, which triggers the trailing head to diffuse, moving it to the leading position, which allows it to bind to tubulin…
That would be interesting.
As we said, the membrane is impermeable to most dissolved solutes. Gases can pass freely through the membrane, so oxygen and CO2 can be traded as necessary. But things dissolved in the water that surrounds the cell and is inside the cell generally are hydrophilic. As such, they cannot pass through the membrane directly.
For this reason, there are transport proteins that are responsible for moving all of the small molecules, ions and even fairly large molecules across the membrane. We will talk a little bit later about the protein that allows water to move across the membrane. For right now we will talk about transport in terms of several overlapping concepts shown in the diagram.

Image from www.physiologyweb.com
Passive diffusion
all forms of passive diffusion are "down gradient." That is to say, one side of the membrane has more of the solute while the other side has less. Given any pathway through which the solute can move it will tend to distribute more evenly. I can divide this further into simple diffusion for molecules that can diffuse again across the membrane directly, passive diffusion for molecules that pass through a channel protein (which is just what it sounds like, a protein that forms a channel through the membrane), and facilitated diffusion, where the solute is carried by a protein from one side of the membrane to the other, sort of like a shuttle. The important thing to remember is that all three of these are diffusion down gradient and require no input energy. No work needs to be done for molecules to diffuse. In fact as we will see later, a gradient can be used to do work.
Active transport.
All forms of active transport require the expenditure of energy at some point in the overall pathway and result in even greater disparities in concentration between the two sides of the membrane. That is, something moves from an area where there is a low concentration to an area where there is already a high concentration of that solute.
Active transport can be separated further into "pumps," which directly expend energy in a cycle to move solutes from one side of the membrane to the other, and "co-transport." Co-transport uses a gradient that was established by a pump or exists for some other reason to drive transport of some other molecule up its own gradient. This is one of those things that is hard to explain in writing. However the diagram below does a pretty good job of showing it.
Symport is when the molecule that is moving down gradient is moving in the same direction as the molecule you want to move up gradient.
This image from your textbook, Campbell Biology ninth edition, and shoes how a proton pump can be used to establish a gradient, then the proton gradient can be used to drive transport of sucrose "up-gradient." This is a symport protien.

In the diagram, we see that ATP is expended to pump protons to the external side of the cell membrane.Those protons, given a pathway, would diffuse down their gradient back into the cell. The trick here is to use a pathway (that is to say, a protein in the membrane) that will only allow protons to diffuse down gradient if a sucrose molecule accompanies it.
You can think of it sort of as a revolving door that will allow protons to enter but will only revolve if sucrose is there also.
The other related concept is the "antiporter." you can think of this as a revolving door where the two players have to enter opposite sides in order for the door to turn. In either case one molecule or ion diffuses down its gradient (passive diffusion) while the other molecule moves up its gradient (active transport).
Ion pumps and the electrochemical gradient
In order to carry out co-transport, one needs to have a gradient established of one or more ions. This is generally achieved by proteins that we call "pumps." While there are many that are crucial to cell function, one has an outsized role in establishing what we call the "electrochemical gradient."That protein is the sodium-potassium pump. Sometimes you will hear it called the sodium-potassium antiporter ( which makes sense because sodium and potassium ions are moving in opposite directions – but is misleading since both will move up gradient against the direction favored by diffusion.) Another name you may hear is the sodium/potassium ATPase.
Here is a moderately amusing GIF based on Drake's music video for "Hotline Bling" from a couple years ago.
Here is the gist of it: the pump can bind two potassium ions when it is open to the outside, but cannot bind sodium ions at all. When open to the inside it will no longer bind potassium ions, but can bind three sodium ions. ATP is hydrolyzed to force the switch between the two states (open to the inside versus open to the outside). The net result of this is that as it goes through a cycle two potassium ions are pumped into the cell and three sodium ions are pumped out. This results in a gradient of potassium (high on the inside, low on the outside) and sodium ions (high on the outside, low on the inside). This is the "chemical" part of the gradient. Because two positive ions are pumped in for every three that are pumped out, there is also an electrical component to the gradient. The inside of the cell ends up being negatively charged overall to a level of about -70 mV.
I would like you to read David G’s really nice blog on the pump. I think you will find some of the numbers, frankly, shocking. Also, understanding the way this works will help you with other proteins we consider.
For example, some may be interested in proton pumps in the cells of the stomach that make the stomach acidic...it works like this protein, as David explains.
Proceeding from specific to general:
The Pump
This protein has binding sites for ions. Below is an image of two ions bound. In this case, the protein crystallographers have used a trick and bound Rubidium+ to the sites. But, it works similarly. What would bind a positive ion? Well, you should hold it between negative ions, like deprotonated acid side chains. What are the acid side chains in amino acids?: Aspartic Acid (D) and Glutamic Acid (E). Look at the image below:

D804, E327 and E779. The numbers correspond to the residue number in the amino acid sequence, or primary structure. So, D804 is an aspartic acid that is the 804th amino acid residue in the chain and is part of one of the helices that span the membrane. E327 is a glutamic acid at position 327, and is in a different helix spanning the membrane, but resting close to D804 in the structure. All of come together to make a really nice pocket that binds the ions (greenish). That makes sense. There are others I didn’t label. All three residues that bind the cation are on different helices. If you slide those helices relative to each other, the components of the binding site move closer or farther away from each other. Now, check out the ionic radii of Na+ and K+ at the wikipedia page. K+ is 30% bigger than Na+ (Rb+ is only slightly larger than K+). So, by sliding the helices, I can change the sized of the pocket and whether K+ or Na+ is favored.
- ATP hydrolysis transfers a phosphate to a serine on the pump (Serine 33 on our pump), causing the helices to shift. This opens the pump to the outside and changes the shape of the binding pocket so that Na+ cannot really be held tightly anymore and K+ fits in well.
- The Binding of K+ changes the shape a little more. This favors hydrolysis of the phosphate from the serine, shifts the helices back to be open inside the cell and makes the pocket too small for K+, but allows Na+ back in. Of course, I didn’t explain exactly, but the three Na+ sites only reconstruct 2 K+ sites once the helices move.
As Goodsell points out, this same mechanism can be used to pump protons. You just have to change the size of the binding pocket. That probably just involves some shifting of the positions of the acidic residues.
Gradients are useful
Once you establish a gradient, you can use it to do stuff. The pump establishes an electrochemical gradient. Every cell at “resting potential” has more K+ on the inside and more Na+ on the outside. The inside is also more negative. Maybe I could use that gradient to do something.
For example:
- Suppose I had a protein that would allow Na+ to come back in...but it would only do that if there was also a glucose molecule bound. I could use the gradient to transport glucose into the cell.
- Suppose I had a channel that I opened in response to some signal...maybe something binding to the channel, and that channel would allow sodium to pass through it. All the sodium ions would start racing into the cell, down their chemical gradient. But that would also make the inside positively charged...that would be a dramatic effect. It get’s even more complicated, but that’s the start of an action potential in nerve cells.
- Or...what if I stacked a whole bunch of cells on top of one another and made a little battery of cells, each with a small voltage across the membrane...then released all of them at once. That would be really interesting.
A more general mechanism for regulated processes
Phosphorylation and interactions with other molecules change the shape of proteins. Changes in the shape of proteins changes their function.
Any protein could have a set of possible shapes, each favored under different conditions, and each favored interaction in turn altering the behavior of the protein.
Imagine a protein that binds to microtubules, for example. Maybe you could come up with a series of shapes that change depending on whether ATP is bound, or ADP is bound, or the protein is bound to tubulin or not. Suppose you had large protein with two heads, each of which can go through a series of steps. Could you imagine a series of steps like: ATP bound to training head, ADP bound to leading head, which binds to tubular, which triggers release of ADP to the solution; which triggers the trailing head to hydrolyze ATP and release phosphate, which triggers the release of the trailing head from tubular, which triggers new ATP to bind to trailing head, which triggers the trailing head to diffuse, moving it to the leading position, which allows it to bind to tubulin…
That would be interesting.