Enolase detail
What is a “mechanism” anyway?
Briefly, a mechanism is a detailed (as detailed as possible) description of the steps of a reaction. You got a little into it last year with the idea of an elemental step in kinetics. However, with the advent of crystal structures of enzymes, more details can emerge.
To review, we have talked about protein structure and the fact that the overall structure results in positioning particular reactive groups of the side chains in a position to carry out chemistry. I’ll add here that metal ions are important factors in the function of many enzymes. Many proteins coordinate metal ions in their active sites and the metal plays an important role in the chemistry (for now, think of “coordinate” as “bind”).
First, some images to get oriented. This is the protein molecule shown in familiar “ribbon” representation.
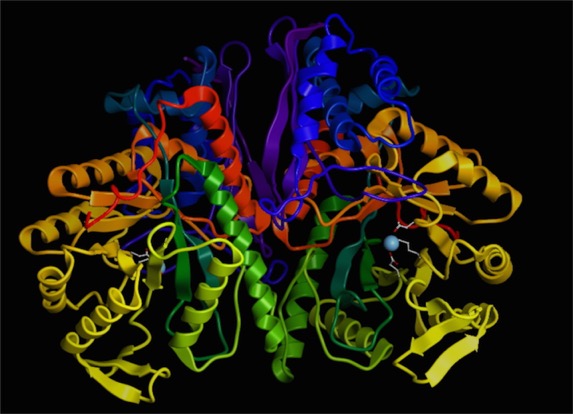
I’ve shown a dimer of two identical proteins. I've only shown the additional detail on the copy on the right. You should see a blue dot, which is the magnesium ion. On the right, you can see a couple of the amino acid residue side chains that interact with the ion. We’ll zoom in on that region here:
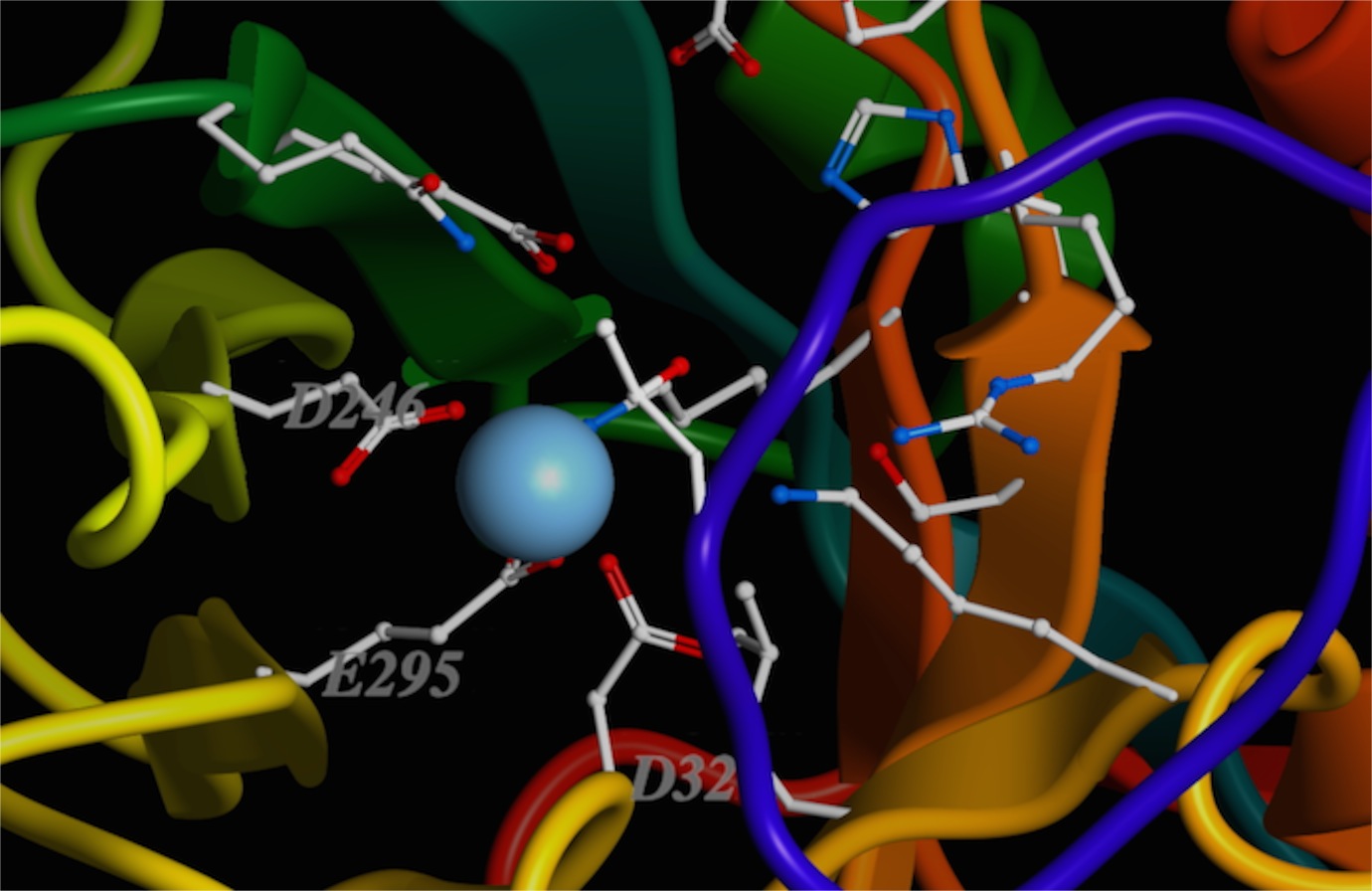
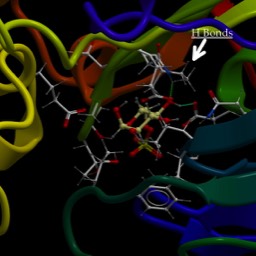
You can see that the positively charged metal ion is being held in place by several side-chain oxygens. I’ve labeled some of them (two aspartic acids, “D,” and one glutamic acid, “E”). There are others. It is probably obvious why the acidic side chains are being used. But, if it isn't, at the pH of the cell, these side chains should be deprotonated, making them negatively charged. Of course, a change to the local pH could change this, couldn't it?
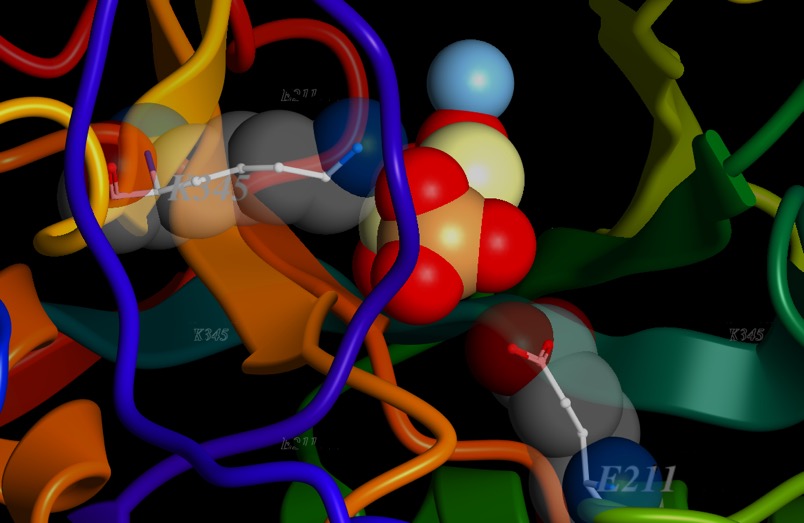
Now, I’m going to add in the substrate, 2-phosphoglycerate. I’ll make it solid, and I’ll highlight a couple of side chains that interact with it, Lysine 345 (K345) and glutamic acid 211 (E211). At the other end of 2-phosphoglycerate, you may see how close the metal ion is to the oxygens of the carboxyl end. The electron-poor magnesium ion is right up against that electron-rich oxygen. Now, normally, oxygen wins the battle for electrons easily...and it does here too. But the whole structure draws electrons away from a bond with a hydrogen at position 2.
Below is a schematic of the reaction of enolase to form PEP, the product of the reaction.
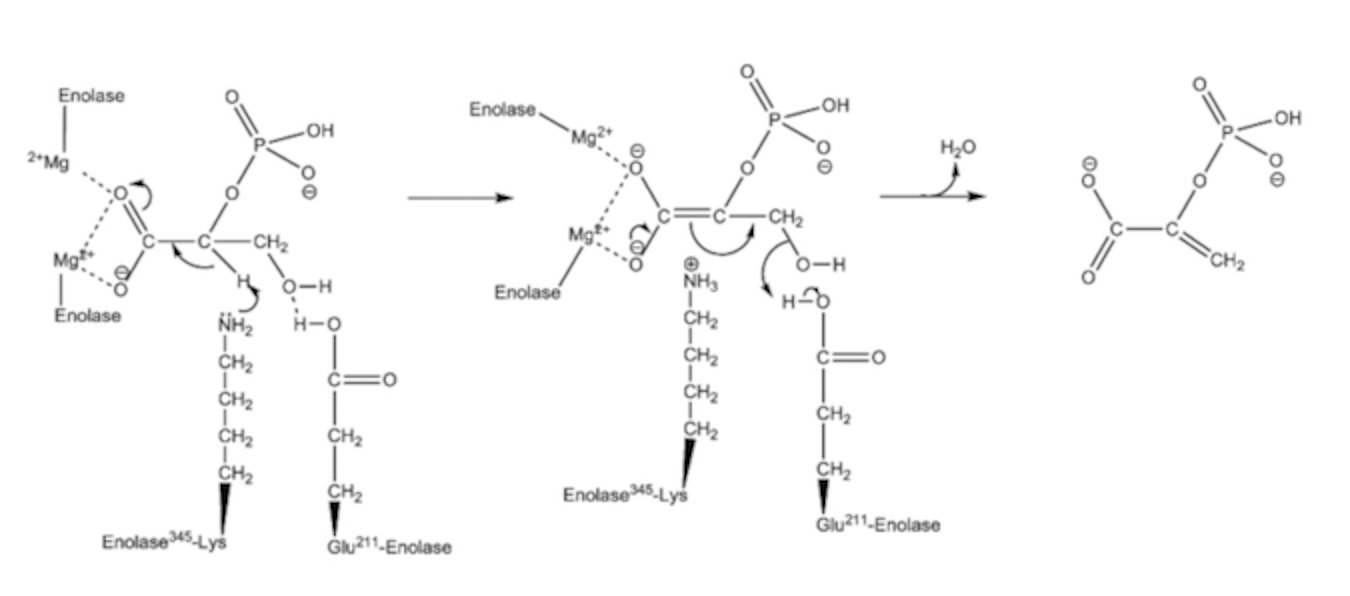
The arrows in the diagram represent moving electron pairs. Here’s what happens:
The metal ion, which is 2+, is withdrawing electrons from the two oxygens, there on the left (this version of enolase has two magnesium ions). Lysine has a basic nitrogen. You see the lone pair depicted on the nitrogen. This makes the H attached to Carbon 2 behave like an acidic proton…not normally the case. However, the proton leaves carbon 2 and gets the electrons in the lone pair on lysine’s nitrogen, the electrons that were in the C-H bond move over and form a double bond with C1. That makes it an “alkene.” The electrons from the double bond of the C=O will flip out to that oxygen, making it negative. The carboxyl of Glu211 is making a hydrogen bond with the OH on C3.
In the second panel, the OH from position 3 will combine with the acidic proton from Glu 211 to form water. The elections from the other carboxyl oxygen will form a double bond, the electrons from the 1-2 double bond will move to the 2-3 position. The product, PEP, is released (Phosphoenolpyruvate). The extra H from the amine of lysine 345 (picked up from carbon 2 in the second panel) transfers over to Glu211 regenerating the enzyme in its starting configuration.
See…kind of cool.