Intro to Enzyme Kinetics
Enzyme Kintetics
Most of the images for this article were taken from the Wikipedia entry on Enzyme kinetics. You can read that here. It gets a little intense, even for our purposes.
Some Words:
Enzyme: a biocatalyst that acts to speed up the rate of a reaction, often by many orders of magnitude (that is, many factors of ten). Most enzymes are protein, though some important ones are RNA.
Catalysts act by lowering the activation energy and, by definition, are not "used up" in a reaction. Thus a catalyst can be used over and over to speed up the same reaction.
Substrate: the starting material acted upon by the enzyme to make the product.
Active site: the site in the enzyme in which the substrate binds. It is the site at which the catalysis takes place.
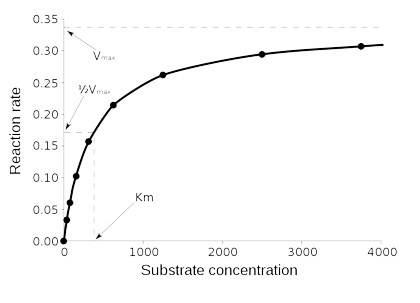
We’ll call the above graph, “fig 1.”
Notice firstly that the graph is determined from a series of kinetic experiments and that Y axis is a rate (usually in mol/L*s that is change in concentration over change in time). So, each point is from an experiment in which you had an initial concentration of substrate and then measured the change in that concentration versus change in time (the initial rate). The points on this Fig.1 are each the initial slope of one experiment from Fig. 2, below.
The image below was taken from Campbell’s website.
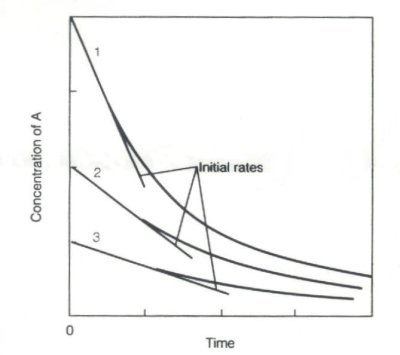 Fig 2
Fig 2It represents three experiments tracking a reaction where A → B and following the rate of disappearance of A. The three experiments have different initial concentrations and different initial rates. The slope of each of those lines becomes a point on Fig 1. The sign of the slope is changed so that it is positive (remember from chemistry that Δ[B]/Δt = -Δ[A]/Δt).
Experiment 1 above might be one of the high points on Fig 1. Experiment 3 would be at lower concentration.
Saturation Kinetics:
The pattern seen in Fig. 1 is called saturation kinetics for reasons that should be obvious. Please Note: the flat part of Fig 1 is not when the net rate of the reaction is zero. It is not equilibrium, but rather the Maximum Velocity (Vmax).
Above a certain concentration of "A," the initial rate no longer increases. The reason for this is that all the enzyme molecules are occupied. Adding more substrate just adds to the “line” of substrate waiting, so to speak (see the “Starbucks Model below). Saturation kinetics are also called “Michaelis-Menten” kinetics after the people who first explained it. You should know that almost no enzymes follow this model exactly. It also deals with a single substrate...but can be adapted to deal with more. Nevertheless, it is an important model that helps explain a lot. The idea is that there are at least two steps to the overall reaction: Binding of substrate to enzyme; and catalysis.
You can think of the process of catalysis as proceeding in at least three steps (there are usually more): a binding step and a catalytic step and a release step. After catalysis, product must be released or the enzyme cannot be reused. Thus, there are at minimum three steps, each with their own rate constants.
Fig.3
“E” is enzyme, “S” is substrate, “ES” is enzyme bound to substrate and “P” is product.
There is an “on” rate and an “off” rate (k1 and k-1) indicating that a substrate molecule can bind and then come off again. The second step is determined by something internal to the enzyme: the catalytic constant. Note that if I blocked the second step, the binding step would come to equilibrium, with substrate binding and releasing at equal rates. Under those conditions, I could calculate a “KD,” for “dissociation constant,” which would be k1/k-1. This would represent the concentration at which you would see ½ maximal binding.
Important Points on Figure 1.
There are two important points on Fig. 1. We have already talked about the first, Vmax. That is, the maximum velocity, or rate, of the reaction once all of the enzyme molecules are busy.
The other point is a substrate concentration known as the Michaelis Constant, or “KM,” which is the concentration at which the rate is ½ of the Vmax. If the catalytic step is fast and we are still in the linear portion of the graph, the KM is approximately equal to the Kd I mentioned above. That is, if the rate of catalysis is very fast compared to the rate of substrate binding, then, E*S is very short-lived and the rate at which product is made is determined essentially by the rate of substrate binding. The book acts as though all enzymes are like this…they are not.
But, when that assumption is correct, the concentration of substrate at which you get half-maximal production of product is the same as the concentration at which you get half-maximal binding of substrate. So, KM≅Kd.
High or Low Substrate concentrations.
Imagine two situations, one in which substrate concentration is low and one in which it is high.
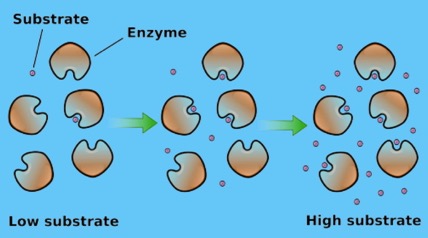 Fig 4.
Fig 4.In the first case, the slow step (rate-determining step) is the binding (first) step. The catalysis step may seem instantaneous, by comparison. This gives us the steep initial portion of Fig. 1. The rate is determined by the rate of binding.
In the latter case, essentially all the enzyme molecules are occupied. No enzymes are sitting around idle. Thus, the reaction is going at its maximum velocity (Vmax), the (nearly) horizontal portion of the graph at the top. At that point, the overall rate is governed by the catalytic step, give by k2.
There is also a k-2. Most enzymes can catalyze the reaction forward or backward. Remember that a catalyst just lowers the activation energy of a reaction and can speed up either direction. The ΔG° does not change.
Finally, product must be released. The product has its own binding constant. Here we have the dissociation of product as k3 and the rebinding of product as k-3.
There are often multiple steps to the catalytic step, each of which could have their own value. In general, we only "see" the effects of the slowest step.
Each of these steps can be affected by binding of other things to the enzyme (allosteric effects). That is, binding of another protein to our enzyme could change its shape a little and change one or more of the ks we have identified. So, the product may bind tightly to the enzyme, which would prevent the enzyme from finding another substrate (this is more common than you might think). But, the binding of another protein may raise the value of k3, which is essentially the dissociation constant for product. That would promote recycling of the enzyme.
Another thing that can change the k values for any step is the addition of a phosphate, as in transfer of a phosphate from ATP to an OH on the side-chain of some amino-acid residue, such as Serine, Threonine or Tyrosine (have I mentioned that hydroxyls…never mind). Since transferring a phosphate back to ADP is generally not favored, this kind of step often seems irreversible (though, not always).
The Starbuck’s Model
Think of the reaction being catalyzed as production of cappuccini at Starbucks. The enzyme is the “Barista” and the customers are the substrate. If you count cappuccini per hour, you get a rate. If there is a low concentration of coffee drinkers wandering in front of the store, you get a slow rate. If you increase their numbers, the rate of cappuccino production increase. The rate is ‘diffusion limited,’ that is, set by the number of coffee drinkers that wander into the store…until a line forms. At that point, the Barista is working at maximum rate (saturation kinetics) and the rate of cappuccino production is limited by the catalytic rate, how fast the Barista can work. The only way to increase the rate is to add more enzyme…in this case, just open another Starbuck’s 100 meters away).
Inhibitors
Let’s consider two types of inhibitors: one that purely inhibits the first step (binding) and one that alters the enzyme so that it is not as good at catalysis (the second step), but otherwise has no effect on binding.
Competitive: Enzyme-Inhibitor (E-I) complex or Enzyme Substrate (E-S)
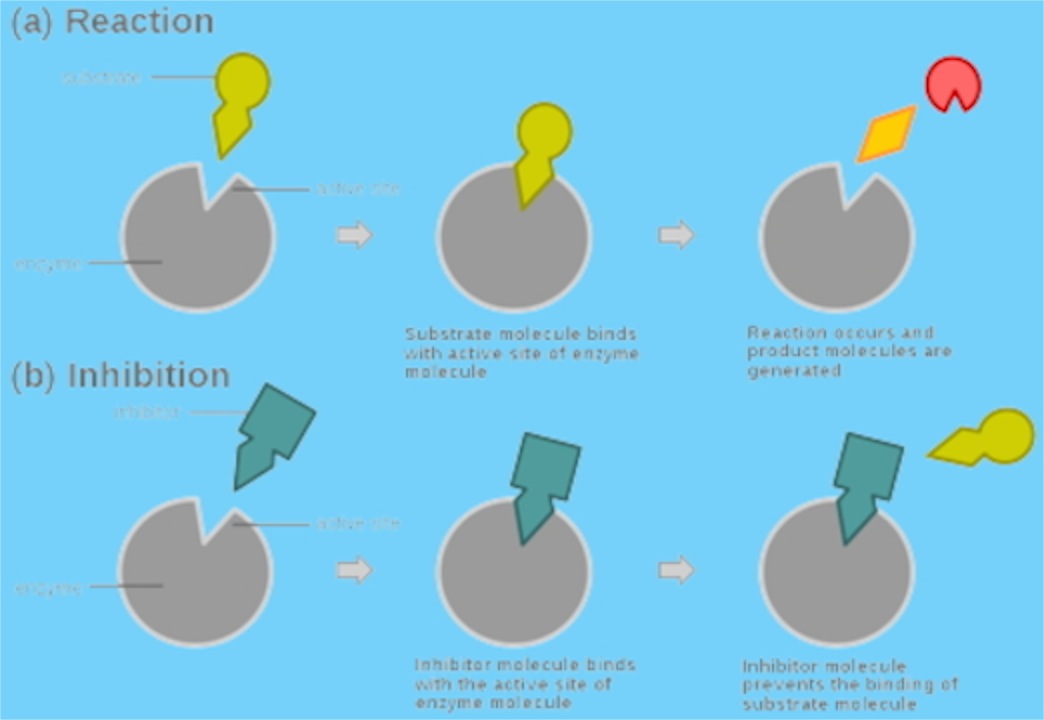 Fig 5.
Fig 5.The substrate does not bind the E-I complex and the inhibitor does not bind the E-S complex. Shown here the two are actually binding to the same site. Inhibitor physically blocks access of the substrate to the enzyme. This could be the idiot who gets to the front of the line at Starbuck’s and cannot think of what he wants to order, but just stands there and won’t let others order.
The substrate will have its binding constant (approximately KM) and the inhibitor with have its binding constant (lets call that KI). Note that the second step governed by k2 in Fig. 3 is NOT affected by the presence of the inhibitor (The Barista works at the same rate…he’s just not able to get working). If the substrate binds, it will be catalyzed at the same rate as it would if the inhibitor were not in solution. It’s just that fewer substrate molecules get to bind.
Since both the inhibitor and the substrate have an “off rate,” I can assume that even in the presence of the inhibitor, substrate will have some opportunity to get access to the enzyme when the inhibitor comes off.
Let’s assume a simple case in which the inhibitor and the substrate have similar binding constants for the enzyme. Then, if I had equal amounts of each, I would expect at any given time that half the enzyme will have substrate bound and half will have inhibitor bound. If I increase the concentration of substrate, I would be able to get more of it to bind (out-compete the inhibitor for the site) and the enzyme would be able to do its job as normal. At a great enough excess substrate concentration, the probability of substrate binding is much higher than that of inhibitor binding; so most enzymes would have substrate bound. I could eventually get the rate up to the Vmax. But, the KM becomes much higher...you need much more substrate to get the enzyme to work at half the max rate.
So, a competitive inhibitor raises the Km, but does not affect the Vmax.
Note: while the mechanism of competition strictly results in inhibition, the other types of interactions described below could just as easily activate the enzyme, or increase, the rate.
Non-competitive: Lower the rate “k2.”
Another type of inhibitor does not affect binding of substrate. Instead, it alters the rate of the catalytic step. How can it do that? Remember that these are proteins. A small shift tertiary structure caused by the binding of inhibitor could change the ability of the enzyme to catalyze the reaction.
This effect where binding of a regulator (inhibitor or activator) changes the shape of the enzyme and alters its rate are call “Allosteric effects.” Allosteric just means binding somewhere else and not directly interfering with substrate binding.
In that case, I’d get binding at the same rate. So, KM would not change. However, since k2 is lower, I have a lower Vmax.
Easy. This would be the girl behind the counter who is really “into” the Barista and is chatting to him non-stop. That slows down his catalytic rate, but doesn’t affect the rate at which customers wander in.
All non-competitive inhibition is allosteric. However, not all allosteric effectors show non-competitive inhibition. Some are even activators.
Regulation
You may notice that I have more steps for my enzyme than the book has. The substrate has a binding constant, what about the product? Wouldn’t I expect the product to have some binding affinity for the enzyme? And, the product has to leave the binding pocket in order for new substrate to fill it. In effect, the product can act as a competitive inhibitor of the enzyme that produced it. This is the very simplest form of “feedback inhibition,” an important concept we will talk more about. We refer to the combined rates for the steps of catalysis and releasing product as an enzyme’s “turnover rate.” That is, how fast it is available to do its job again.
Much of the time, I don’t want an enzyme to work at full speed. One very important way of regulating an enzyme is feedback inhibition, as in the simple product competitive inhibition mentioned above. Suppose I have a pathway requiring 3 enzyme steps.
Once I have enough D, I don’t want the pathway to be running at full speed. Maybe “A” is also needed for another pathway (not at all unlikely) and once I have enough D, I want to divert A down that other pathway.
One real world example would be if "A" were glucose. Sometimes I want to use my glucose to get energy right now. Sometimes I want to store glucose in glycogen for later use. Which path I choose would depend on changing conditions. A well-evolved system should adjust to needs.
One way to regulate this is if D is an inhibitor of enzyme 1. It may be competitive or non-competitive.
Allosteric activation is common as well, as mentioned above.
This sounds magical, until you remember that we are talking about proteins, composed of helices and sheets arranged in specific tertiary structure. Couldn't the binding of some activator on one face of the protein cause the helices to shift relative to each other? Of course. Couldn't the shift slide a helix out of the way of the active site, allowing better access? Sure.
The possibilities for regulation seem endless, really. And every possibility I've ever thought of and about 100x more that I never thought of can be found operating in nature.