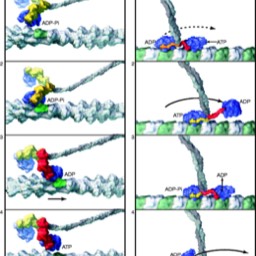
Nucleic Acids
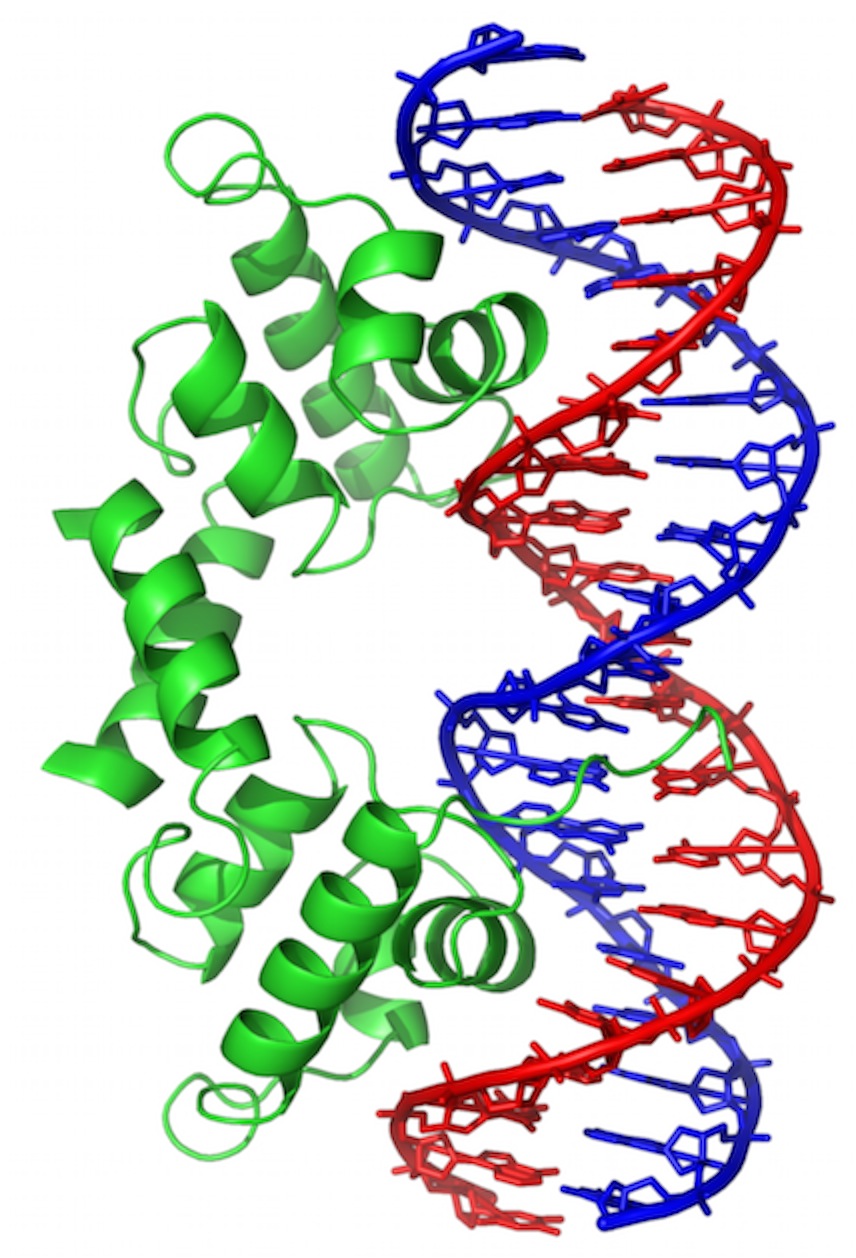
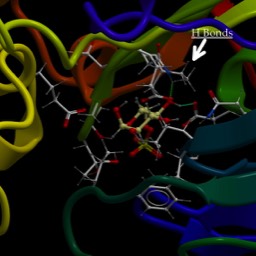
Wikipedia wikimedia commons image of a Short stretch of DNA bound by a protein called “Lambda Repressor.” DNA bases are shown in “stick model” form, sugar-phosphate backbone as red or blue pipes. Protein is shown in “ribbon” form, backbone only (no sidechains).
This is one of the ways that genes are turned “off” and “on.” In this case, it’s for a gene found in a virus that infects bacteria. But, many of the same principles will apply all over.
On Friday, you will be doing a model-building lab. Please be careful with my model. You will follow the instructions in this document.
The Double Helix
This is a really beautiful structure. Unfortunately, the beauty lies in just how much it explained when it was first proposed, and is therefore, perhaps, hard for you to see. To me, it is astonishing. I’ll run down a few features.
Basic Structure (image also from Wikimedia Commons):
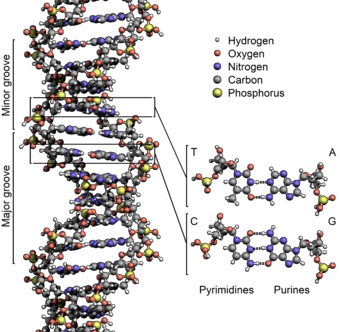
And a more detailed view:
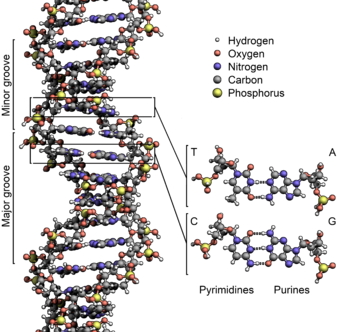
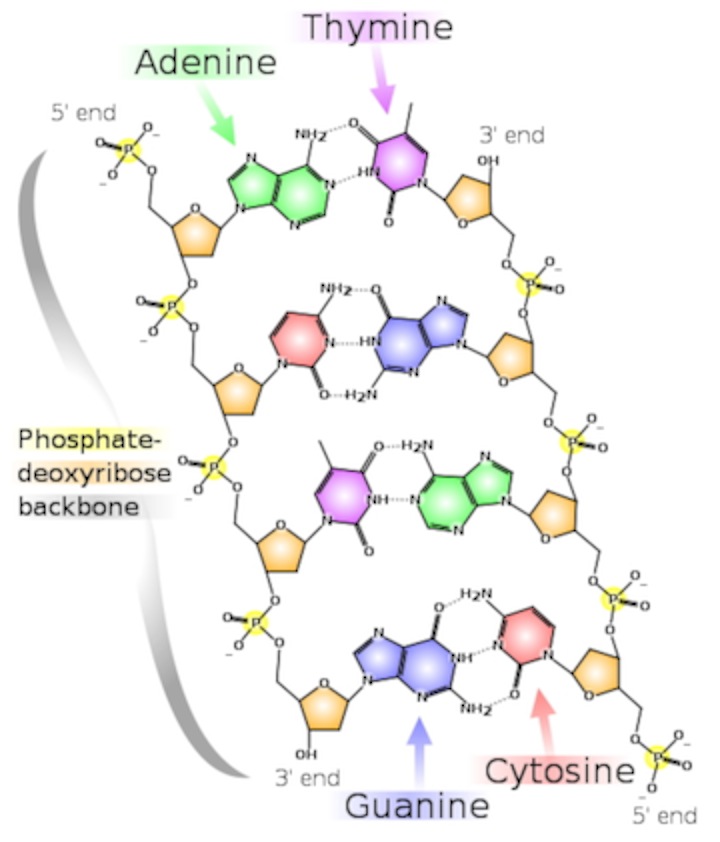
- There are two strands. Each is defined by a polymer of sugar-phosphate moieties (yes, that’s a word similar in meaning to “functional group”).
- We number the carbons in the rings. Ribose (and deoxyribose) are pentoses, or five-carbon sugars. In this structure, the number 1 carbon has the "base" (A, T, G or C) attached to it. The number 5 carbon is outside the ring. Each subunit is linked from the number 3 carbon of one deoxyribose, to a phosphate, then to the number 5 carbon of the next deoxyribose. Instead of calling them 3 and 5, we call them 3’ (pronounced 3-prime) and 5’. The “prime” tells you that the number refers to the carbons in the sugar, as opposed to a carbon in the rings of the “base.”
- Thus, each strand has a chemical polarity. What I mean is that each strand has a 5’ end and a 3’ end, just as protein has an amino end and a carboxyl end. That, it turns out, is not a coincidence.
- The strands are aligned “anti-parallel.” The 5’ end of one strand of the double helix is aligned with the 3’ end of the other.
- The information is encoded by the “bases,” which protrude from the polymer backbone. This is really one of the key points: DNA is a code. The sequence of bases stands for something else. The main thing a DNA sequence stands for is the sequence of amino acids in a protein. A good first definition of “Gene” is: a stretch of DNA that encodes a protein.
- The information takes the form of hydrogen bonds. This is more profound than it seems: information is energy. Adenine has a much greater ability to bond to Thymine than to Cytosine; Guanine bonds much more tightly cytosine. Thus, we have A-T pairs and G-C pairs. Each base “complements” the one it pairs with in terms of hydrogen bonds it can make. We use the term “complementarity” to describe this.
- Now, for some interesting stuff: DNA is redundant. I only have to see one strand to know what the sequence of the other is. Each stand has all the information needed to specify its opposite strand. When there is damage to one strand, the other strand tells you how to fix it. To replicate, each strand directs the synthesis of its mate.
- The information is in the hydrogen bonds...and it’s a code that specifies the primary sequence of proteins (among other things). Yes, I am aware that I too am being redundant. It’s important. The DNA and the peptide sequence are “co-linear.” The 5’ end of a gene corresponds to the amino end of the peptide chain. The 3’ end of the gene corresponds to the carboxyl end of the protein. Each group of three bases in a gene is a “codon,” specifying what amino acid goes in the corresponding place of the protein...Obviously, some fascinating machine and lots of regulation must go into that. I could go on and on...but I won’t just yet.
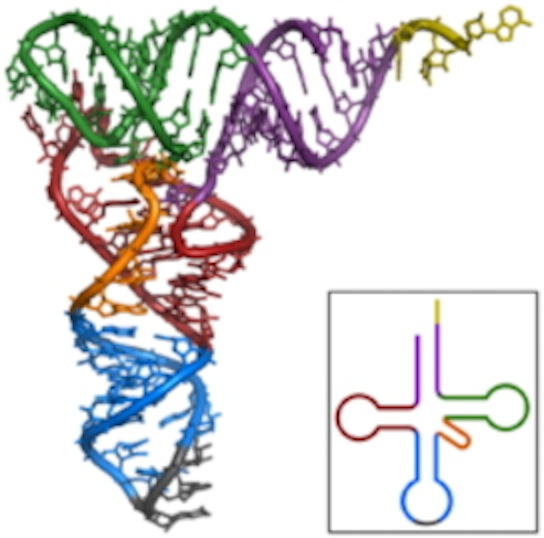
RNA
RNA
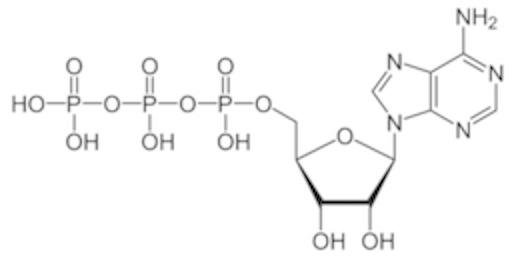
- This is ATP (adenosine triphosphate). You’ve heard of it in terms of an “energy source” for your body. That’s true, as far as it goes. However, It is also one of the four bases that are the building blocks of RNA. The difference between this and dATP (“d” for “deoxy”) is the presence of that oxygen on the 2’ carbon. (Note, when I draw it, I usually have the protons removed from the phosphates. It’s an acid and will deprotonate under most conditions in the cell. this diagram I lifted happens to have the phosphates protonated).
- Here are three important differences between RNA and DNA:
- There is the extra oxygen.
- RNA is almost always single stranded, but that strand can fold back to form stretches of helix. Remember that if you fold a strand back, the arrangement is antiparallel, which is the way nucleic acids align when pairing.
- The rules for base-paring are the same…except that a different pyrimidine, “Uracil” or “U” stands in for Thymine. It’s pairing is no different. It just lacks a methyl group on the pyrimidine ring.